Group Versteeg
Nuclear degradation mechanisms of Activation Induced Deaminase
Group Leader
Gijs Versteeg
Gus Versteeg started his independent research group at the Max Perutz Labs Vienna in 2013. His previous work explored how ubiquitination mediates immune system activation. His group currently investigates how the abundance of immune- and cancer-related proteins is controlled by proteasomal degradation.
- Institute Max Perutz Labs, Univ. of Vienna
- Phone +43 1 4277 54637
- Mail gijs.versteeg@univie.ac.at
- Web https://www.maxperutzlabs.ac.at/versteeg
Projects within consortium
Activation Induced Deaminase (AID) controls class-switch recombination and somatic hypermutation in B cells. AID is associated with cancer hyper-mutation, and degraded in a nuclear, proteasome-dependent manner. We aim to identify cellular factors mediating AID degradation, and determine how this is mechanistically achieved. Characterizing the factors that mediate AID turn-over will provide insight into cellular ‘genome guardian’ mechanisms that prevent DNA damage.
The APOBEC3-related Activation Induced Deaminase (AID) controls class-switch recombination and somatic hypermutation in B cells. Like APOBEC 3A/B/H-I, AID is associated with cancer hyper-mutation, and degraded in a nuclear, proteasome-dependent manner.
We hypothesize that there is a nuclear proteasome-dependent AID degradation pathway that limits accumulation of genotoxic AID. In this study, we aim to address the following questions: i) what are the factors that control AID protein abundance in B cells?, ii) how do ubiquitin-dependent and -independent processes contribute to AID degradation?, iii) do specific AID physicochemical properties make it a proteasome substrate, or is it pre-processed by other cellular components?, and iv) how important is nuclear AID degradation for restricting genome aberrations?
Identifying this AID-targeting axis will provide key insights into how cells target AID and how this is mechanistically achieved. Therefore, results from this project have the potential to provide novel understanding of the players and mechanisms by which this is achieved in ubiquitin-dependent and -independent ways.
Understanding the factors that limit AID accumulation, and their role in preventing hyper-mutation, will provide valuable insights in to the cross-deaminase-family mechanisms that evolved to enable proper antiviral and immune-regulatory activities on the one hand, yet prevent host-detrimental effects on the other is a major driver of mutation in various cancers.
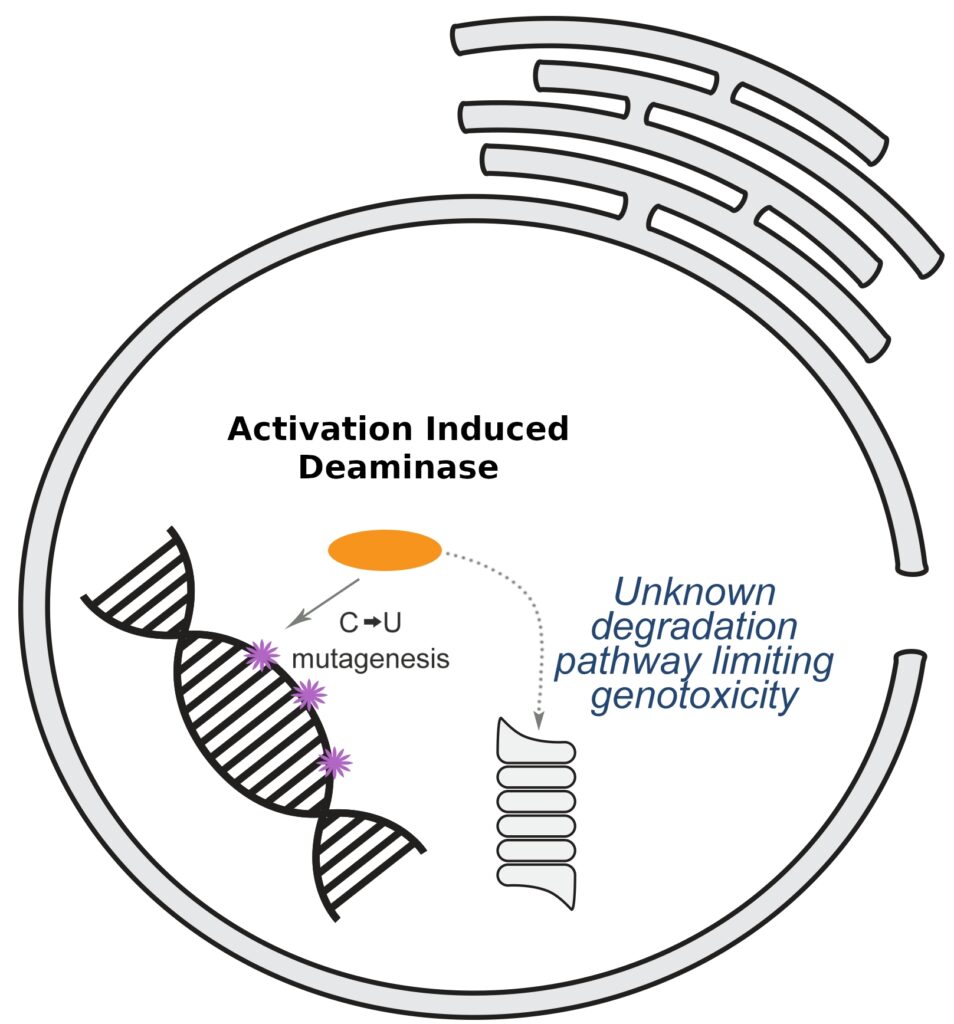
Activation Induced Deaminase (AID) is essential for physiological genome editing in B cells. This project aims to identify factors degrading nuclear AID, thereby preventing aberrant host genome mutagenesis.
Project members
-
PhD Student
Irene Schwartz
SFB Member
-
PhD Student
Alexandra Shulkina
SFB Member
-
PhD Student
Valentina Budroni
Associated
Targeted Protein Degradation related publications by Group Versteeg
- 2025 TRIM52 maintains cellular fitness and is under tight proteolytic control by multiple giant E3 ligases Nature Communications Go to publication →
- 2024 Disordered regions in the IRE1α ER lumenal domain mediate its stress-induced clustering EMBO Journal Go to publication →
- 2024 Guardian ubiquitin E3 ligases target cancer-associated APOBEC3 deaminases for degradation to promote human genome integrity bioRxiv Go to publication →
- 2023 SPOP targets the immune transcription factor IRF1 for proteasomal degradation eLife Go to publication →
- 2023 HUWE1 controls tristetraprolin proteasomal degradation by regulating its phosphorylation eLife Go to publication →
- 2023 Structural basis of how the BIRC6/SMAC complex regulates apoptosis and autophagy Science Go to publication →