Group Clausen
Spatial control of E3 ligases
Group Leader
Tim Clausen
Tim Clausen conducted his PhD studies in 1994 at the at the MPI of Biochemistry in Martinsried and continued there as postdoc in 1997. In 2002, he became Group Leader at the IMP in Vienna, where he is now Senior Group Leader since 2009.
- Institute IMP - Research Institute of Molecular Pathology
- Phone +43 1 79730 3300
- Mail tim.clausen@imp.ac.at
- Web https://www.imp.ac.at/groups/tim-clausen/
Projects within consortium
Our research focuses on understanding how the cellular localization of ubiquitin ligases affects their substrate targeting. To address the molecular GPS mechanism, we will explore how HAPSTR1 controls the shuttling of HUWE1 between nucleus and cytosol and, in parallel, examine the effect of post-translational modifications on RNF213, directing it to lipid droplets to target microbial invaders.
Our group is interested in molecular mechanisms underlying the spatial and temporal control of ubiquitin ligases. One of our models, HUWE1, is a highly conserved ubiquitin ligase that plays an important role in protein quality control, targeting unassembled orphan proteins. Our second model, RNF213, is a first-line defender in the innate immune response, marking invading pathogens for subsequent degradation. How the substrate targeting and activity of the two giant E3 ligases is regulated in the cell is not known.
To address this point, we aim to identify and characterize specific co-factors of HUWE1 adjusting its promiscuous E3 activity towards specific substrates. Using an integrative structural and cell biology approach we seek to characterize the interplay between HUWE1 with identified co-factors, including a specialized chaperone and HAPSTR1. Our current data point to a tightly controlled mechanism of how HUWE1 transits between different cellular compartments, linking compartmentalization of E3 ligases with their functional control.
For RNF213, we aim to understand its specificity for targeting bacterial lipids and the role of lipid droplets within this process. In fact, RNF213 was the first ubiquitin ligase shown to ubiquitinate non-protein substrates, opening a new chapter in ubiquitin biology – an exciting yet technically challenging area at the interface of the ubiquitin-proteasome system and biological membranes. Using novel chemical biology tools we seek to explore this uncharted territory, aiming to reveal the role of lipid droplets as RNF213 distribution centers and the molecular mechanism used to ubiquitinate specific lipid molecules.
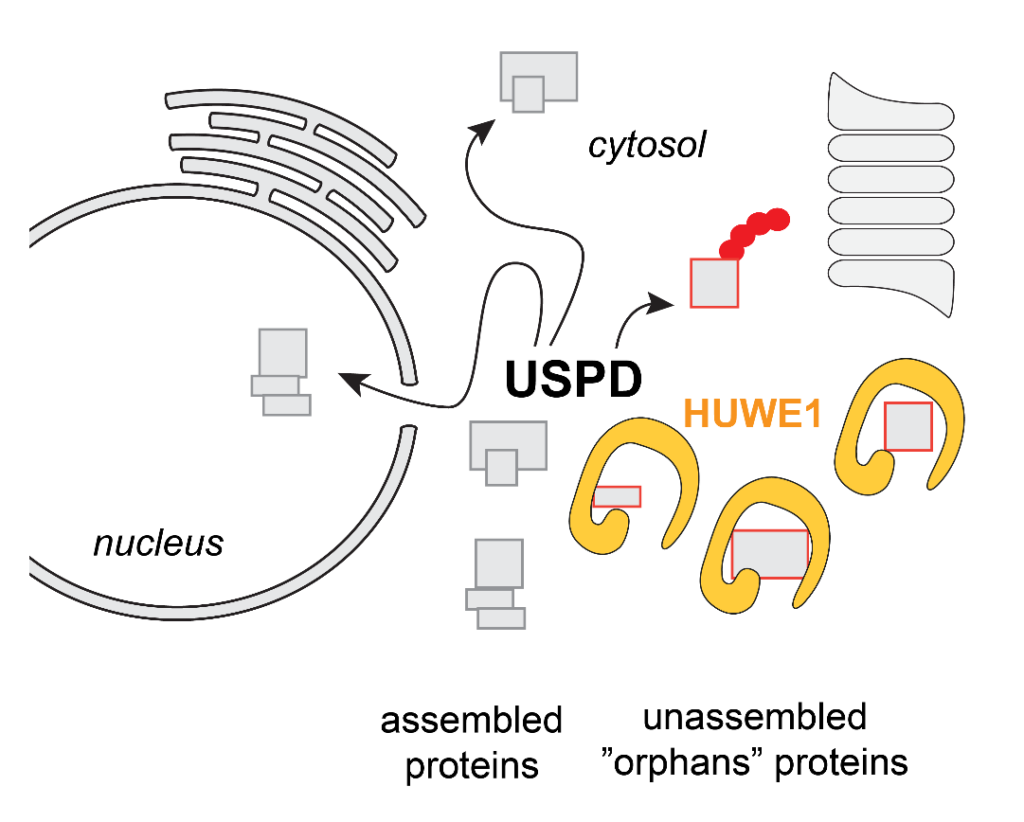
The HECT E3 ligase HUWE1 is part of the USPD (Unassembled Soluble Protein Degradation) pathway, marking orphan protein for proteasomal degradation. Among its >100 client proteins are various transcription factors as well as isolated histone and ribosomal subunits.
Project members
-
PhD Student
Victoria Faas
SFB Member
-
PhD Student
Rebeca Gorgova
SFB Member
Targeted Protein Degradation related publications by Group Clausen
- 2025 TRIM52 maintains cellular fitness and is under tight proteolytic control by multiple giant E3 ligases Nature Communications Go to publication →
- 2024 Cross-species interactome analysis uncovers a conserved selective autophagy mechanism for protein quality control in plants bioRxiv Go to publication →
- 2024 Guardian ubiquitin E3 ligases target cancer-associated APOBEC3 deaminases for degradation to promote human genome integrity bioRxiv Go to publication →
- 2023 HUWE1 controls tristetraprolin proteasomal degradation by regulating its phosphorylation eLife Go to publication →
- 2023 Structural basis of how the BIRC6/SMAC complex regulates apoptosis and autophagy Science Go to publication →
- 2023 Shuffled ATG8 interacting motifs form an ancestral bridge between UFMylation and C53-mediated autophagy EMBO Journal Go to publication →
- 2021 E3 ubiquitin ligase RNF213 employs a non-canonical zinc finger active site and is allosterically regulated by ATP bioRxiv Go to publication →
- 2021 HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain Nature Chemical Biology Go to publication →