Previously a member: Group Loizou
Nuclear quality control and DNA damage
Former Group Leader in this consortium
Joanna Loizou
Joanna Loizou completed her undergraduate studies and PhD in the UK. Postdoctoral work followed at the IARC, WHO, France and the London Research Institute, Cancer Research UK. Joanna established her independent group in 2011 in Vienna at CeMM and became professor in cancer biology at Medical University of Vienna in 2020.
Joanna Loizou closed her project within this program in August 2022 as she moved from Vienna to UK.
- Institute formely: CeMM & MedUni Vienna
- Phone n/a
- Mail n/a
- Web https://cemm.at/research/former-groups/joanna-i-loizou
Projects within consortium
The human genome is constantly exposed to endogenous and exogenous sources of DNA damage. DNA repair ensures the integrity of large eukaryotic genomes by minimising the mutation rate. Several highly effective pathways for DNA repair with distinct specificities exist, which are evolutionary conserved despite partial redundancies. Our lab investigates how cellular decisions are made to engage one repair pathway over another and the molecular mechanisms that lead to DNA repair.
Helix distorting DNA damage, that is encountered by RNAPII, is mostly repaired by transcription-coupled nucleotide excision repair (TC-NER). When TC-NER cannot allow for continued transcription, the stalled RNAPII is removed to allow for access by DNA repair proteins that function to repair the DNA lesion. Removal is thought to be achieved by poly-ubiquitination and degradation of RPB1, the largest subunit of RNAPII. Hence, degradation of RPB1 following DNA damage functions to regulate two fundamental cellular process: DNA repair and transcription.
Our overall goal is to understand how the UPS interacts with RNAPII to clear stalled transcription complexes from sites of DNA damage, in human cells. Firstly, we will identify genetic factors required for (de)stabilization of RPB1. To do so we will use a focused sgRNA CRISPR library targeting E3 proteins, deubiquitylating (DUB) enzymes, and accessory proteins (that function within this pathway), hence identifing which proteins (de)stabilize RPB1. Secondly, we will identify potential E3 ligases and DUB enzymes that add or remove poly-ubiquitin from RPB1, taking orthogonal proteomics-based approaches. Finally, we will investigate the mechanisms by which the identified E3s and DUBs affect RPB1 degradation and stabilization, as well as the subsequent cellular outcomes. Here, we will map the type and position of ubiquitylation mediated by the identified E3 and removed by the DUB. We will determine the impact of ubiquitylation/deubiquitylation of RPB1 on DNA repair and transcription.
Taken together, this multidisciplinary proposal will clarify mechanisms and outcomes of RNAPII degradation following DNA damage, in order to coordinate transcription and DNA repair.
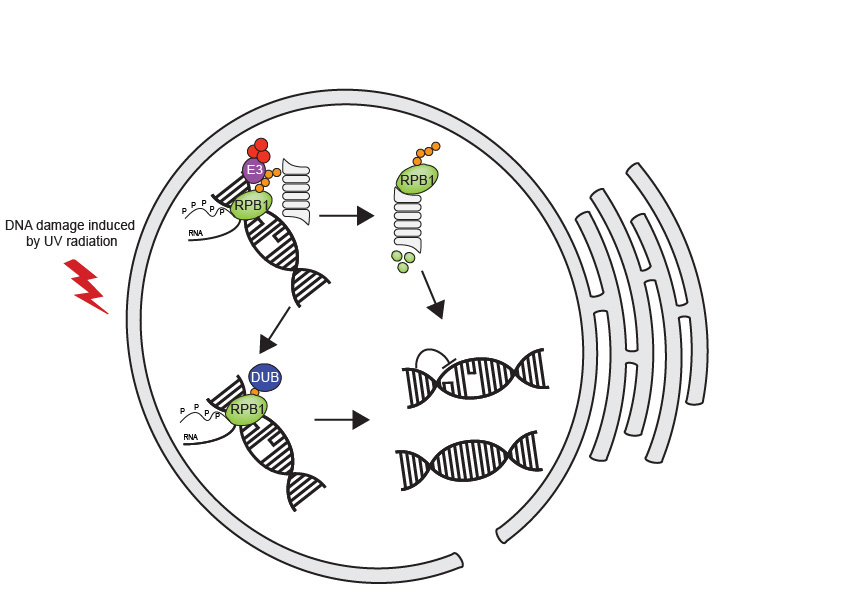
Our goals are to identify the E3 ligases and accessory proteins that degrade RPB1, the DUBs that can cleave ubiquitinated RPB1 and the effects on transcription and DNA repair.
Project members
-
Former SFB Postdoc
Sara Bernardo
(2020-2022)
Targeted Protein Degradation related publications by Previously a member: Group Loizou
- 2022 POLθ processes ssDNA gaps and promotes replication fork progression in BRCA1 deficient cells Cell Reports Go to publication →
- 2021 Decomposing the mutational landscape of cancer genomes with RepairSig Cell Systems Go to publication →