Group Haselbach
Ubiquitin chain recognition and editing by the human 26S proteasome
Group Leader
David Haselbach
David Haselbach finished his PhD in Biophysics at the MPI for Biophysical Chemistry. After a postdoctoral stay in the same institute he became Fellow for electron microscopy at the IMP in Vienna in 2017 and became Group Leader there in 2020.
- Institute IMP - Research Institute of Molecular Pathology
- Phone +43 1 79730 3009
- Mail david.haselbach@imp.ac.at
- Web https://www.imp.ac.at/groups/david-haselbach/
Projects within consortium
Proteasomal degradation serves as key event in the regulation of a plethora of cellular processes. While the general mechanism of proteasomal degradation is well established, key events such as the ubiquitin chain binding and processing as well as the product release are not. The Haselbach lab seeks to address these questions with modern cryo EM and biophysical methods.
Cell division and differentiation depend on the proteasomal degradation of regulating proteins like securin or myc. However, how the proteasome recognizes and distinguishes substrates is still largely unknown. Prerequisite for degradation of a target is its posttranslational modification with a ubiquitin chain. However, ubiquitin is a versatile signal. Summarized as the ubiquitin code, thousands of diverse polyubiquitin chain topologies can be formed with different lengths and linkages. However, how the proteasome selectively distinguishes between these topologies is unclear, as the early substrate-binding and initiation steps have previously been invisible to conventional approaches. This is due to the efficient rate of catalysis by the proteasome and additional editing of ubiquitin chains by proteasome associated factors. Using innovative approaches, including time-resolved cryo-electron microscopy (trEM) sample preparation, deep learning based image processing to extract thermodynamic insights, in-situ cryo EM and single-molecule techniques we will examine a systematic set of polyubiquitin chains. We will employ these approaches first to apo-proteasome complexes but will later extend to proteasomes that contain ubiquitin chain editing enzymes as they would be found in the cell.
On a broader perspective the Haselbach lab is interested in the proteasomes regulation and adaptation in stress and disease states.
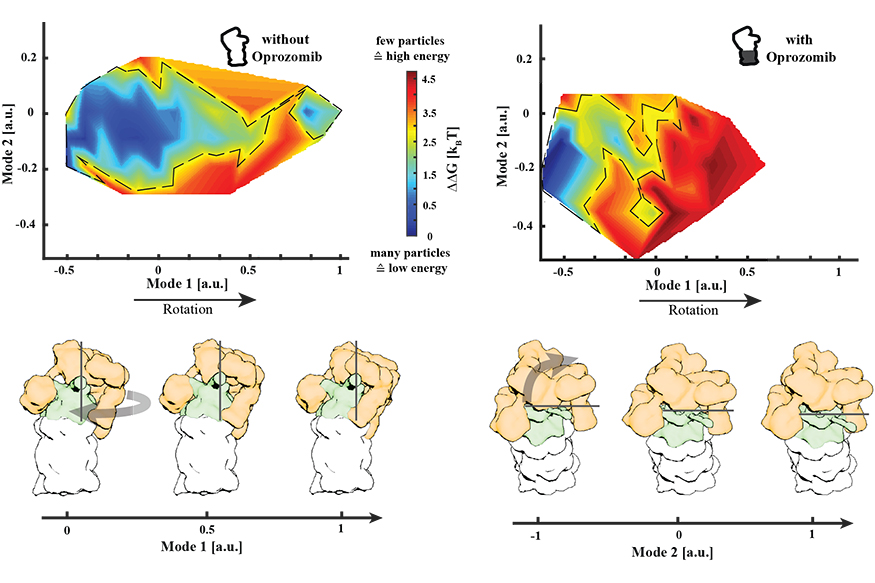
Change of the conformational landscape of the proteasome upon inhibitor binding. Energy landscapes with and without the drug Oprozomib are depicted. Without Oprozomib, the energy landscape is rather broad and flat which allows proteasomes to sample a wide range of conformations without facing a significant energy barrier. In contrast, upon drug binding, well-separated minima can be observed next to a significant energy barrier (red) that restricts the conformational space that can be sampled by the proteasome. A 3.8 Ångstrom resolution structure was determined from particles belonging to this proteasome conformation in a local energy minimum (dark blue). A graphical representation of the movement modes is displayed below.
Project members
-
PhD Student
Hannah Knaudt
SFB Member
Targeted Protein Degradation related publications by Group Haselbach
- 2025 The UBR domain of plant Ubr1 homolog PRT6 accommodates basic and hydrophobic amino termini for substrate recognition Journal of Molecular Biology Go to publication →
- 2024 PITHD1: An Endogenous Inhibitor of the 26S Proteasome During Cellular Dormancy bioRxiv Go to publication →
- 2024 A multivalent adaptor mechanism drives the nuclear import of proteasomes bioRxiv Go to publication →
- 2024 Electrostatic changes enabled the diversification of an exocyst subunit via protein complex escape bioRxiv Go to publication →
- 2024 Guardian ubiquitin E3 ligases target cancer-associated APOBEC3 deaminases for degradation to promote human genome integrity bioRxiv Go to publication →
- 2021 HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain Nature Chemical Biology Go to publication →