Group Becker
Tailor-made ubiquitin chains for targeting the UPS or autophagy
Group Leader
Christian Becker
Christian F. W. Becker obtained his PhD in Chemistry in 2001. After becoming a group leader at the Max-Planck Institute of Molecular Physiology in 2004 and associated professor at the TU Munich in 2007, in 2011 he accepted a call to the University of Vienna as Full Professor and Head of the Institute of Biological Chemistry.
- Institute Institute of Biological Chemistry, University of Vienna
- Phone +43 1 4277 70510
- Mail christian.becker@univie.ac.at
- Web https://biologischechemie.univie.ac.at/en/research/becker/
Projects within consortium
Access to homogeneous, ubiquitinated proteins provides a unique tool in deciphering the complex ubiquitin-proteasome system (UPS) and the autophagy-lysosomal pathway (ALP), as the exact molecular composition of ubiquitin-tags is critical for recognition by parts of the UPS and ALP. To provide such tools, we will generate a set of modified Ub building blocks via protein semisynthesis by combining solid phase peptide synthesis (SPPS) and expression of Ub-intein fusion constructs in E. coli.
The complexity of targeting substrates to the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal pathway (ALP) is based on several aspects, such as the 8 different linkage types possible in ubiquitin (Ub) chains, the chain length, the modification site(s) in the target proteins and, only recently discovered, PTMs of Ub. Several approaches to assemble homogeneous, mono- and polyubiquitinated peptides and proteins that cannot easily be isolated from natural sources have been developed over the past three decades. Among them, enzymatic approaches that have advanced to a stage where different linkages and modification sites can be addressed if E2 and E3 enzymes are available but control over chain length and overall homogeneity is still lagging behind.
Combining intein-based generation of activated Ub chains with solid phase peptide synthesis (SPPS) to obtain Ubs with a potentially large set of modifications has arguably been the most successful strategy to generate fully defined Ub chains and Ub-modified proteins. As defined Ub chains can now be site-selectively linked to target proteins, degradation signals can be deciphered at much higher molecular resolution as demonstrated for several target proteins. In combination with defined Ub chains and binding assays for key components of the UPS and ALP, we will investigate the role of site-specific attachment of differently linked chains of up to 4 Ubs within this SFB.
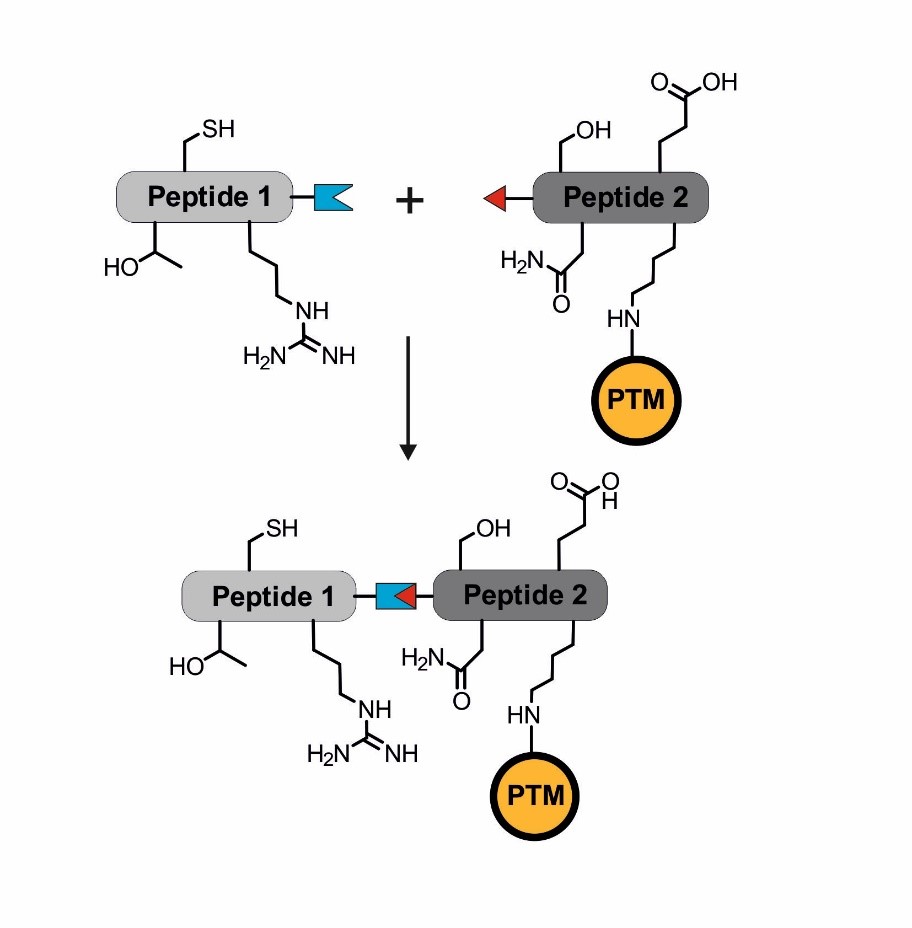
Introducing posttranslational modifications via protein (semi-)synthesis. The blue and red linkage indicates the site of a chemoselective ligation to combine two peptide segments that can be of synthetic or biological origin.
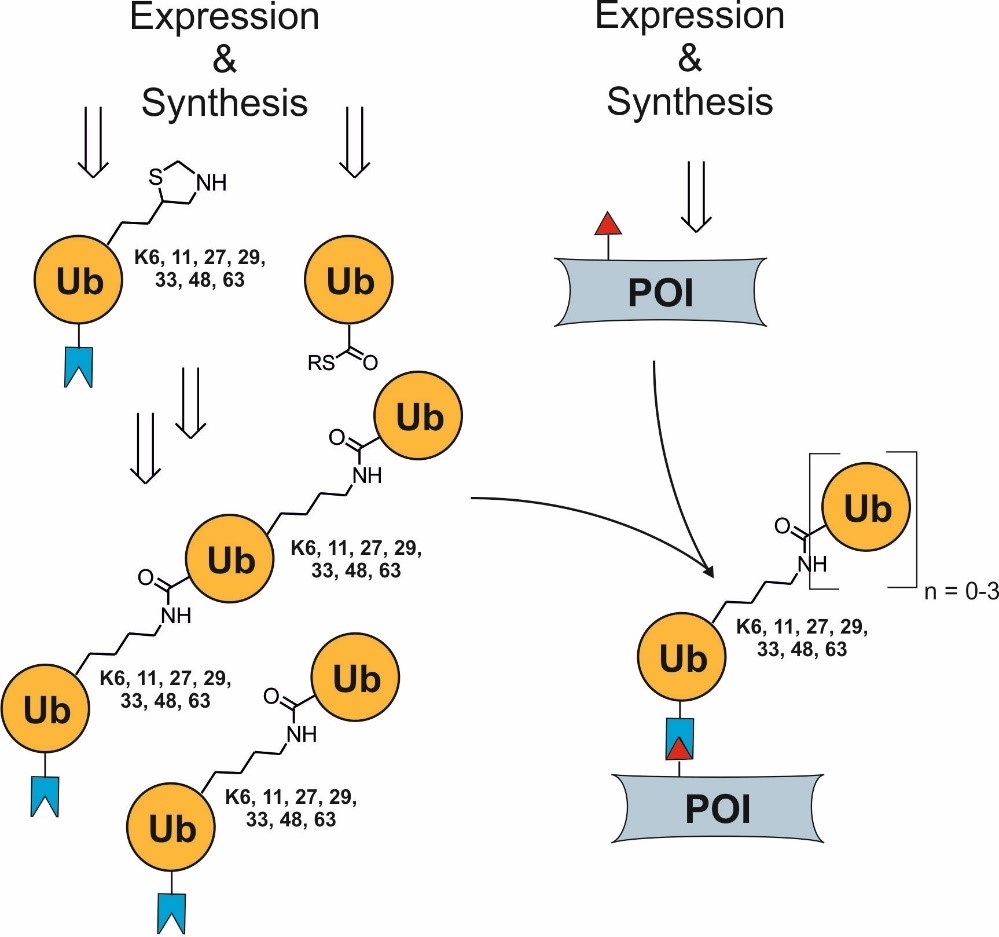
Assembly of homo- or heterotypic ubiquitin chains of different but fully controlled length using chemoselective ligation reactions.
Project members
-
Postdoc
Moritz Urschbach
SFB Member
-
Former SFB Postdoc
Susanne Huhmann
(until 2022)
-
Technical Assistant
Gerhard Niederacher
Associated
-
PhD Student
Dominik Vogl
Associated
Targeted Protein Degradation related publications by Group Becker
- 2025 Convergent Assembly of Homo- and Heterotypic Ubiquitin Chains from Functionalized, Expressed Monomers via Thiol-Ene Chemistry Angew Chem Int Ed Engl Go to publication →
- 2023 Light-Cleavable Auxiliary for Diselenide–Selenoester Ligations of Peptides and Proteins Chemistry Europe Go to publication →
- 2022 Site-selective photocatalytic functionalization of peptides and proteins at selenocysteine Nature Communications Go to publication →
- 2022 Expressed Protein Selenoester Ligation Angewandte Chemie International Ed. Go to publication →