Group Winter
Molecular glue degraders: chemical probing and functional characterization of DCAFs
Group Leader
Georg Winter
Georg Winter obtained his PhD from the Medical University of Vienna under the supervision of Giulio Superti-Furga, and went on as a postdoc with James Bradner at the Dana Farber Cancer Institute/Harvard Medical School. Georg was recruited as a CeMM Principal Investigator in 2016 where his research is now focused on using the unique molecular pharmacology of targeted protein degradation.
- Institute CeMM - Research Center for Molecular Medicine
- Phone +43 40160 70031
- Mail gwinter@cemm.oeaw.ac.at
- Web https://www.winter-lab.com/
Projects within consortium
Our lab is fascinated by research at the interface of chemical biology, gene regulation, proteolysis and cancer. In my research laboratory, we want to develop and apply novel chemical tools to probe and understand biological processes at a high kinetic resolution. In all aspects of our research, we aim to describe and comprehend research questions on a systems-wide level. We are deploying an integrative research strategy to understand how transcriptional networks are rewired in order to sustain an oncogenic state and how we could target resulting dependencies on the transcriptional machinery or associated chromatin regulators. In particular, we are trying to further accelerate the emerging field of targeted protein degradation and leverage its unique molecular pharmacology to elevate our understanding of oncogenic gene regulation and proteostasis.
Here, we propose a multi-disciplinary approach to chemically probe DCAFs to identify novel small molecular glues capable of re-directing their activity. Towards that end, we will deploy a scalable screening system to identify small-molecules capable of binding to DCAFs in cells. This will be followed by a target identification campaign supported by quantitative proteomics and chemical genetics strategies. Finally, we aim to elucidate structural determinants for the drug-induced molecular recognition of neo-substrates by deciphering the underpinning degron sequence and assaying kinetic determinants of ternary complex formation. If successful, this proposal will unlock novel corners of the “undruggable proteome” by identifying novel molecular glues to hijack the activity of DCAFs to degrade proteins outside the scope of traditional ligand-discovery efforts.
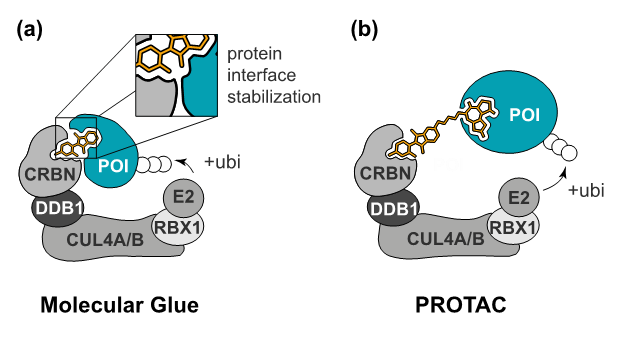
Two different strategies to induce targeted protein degradation. (a) Molecular glues are monovalent compounds that induce the dimerization of two proteins, here an E3 ligase substrate receptor and a neo-substrate. (b) PROTACs are heterobifunctional degraders that can individually bind to the E3 ligase and the protein of interest.
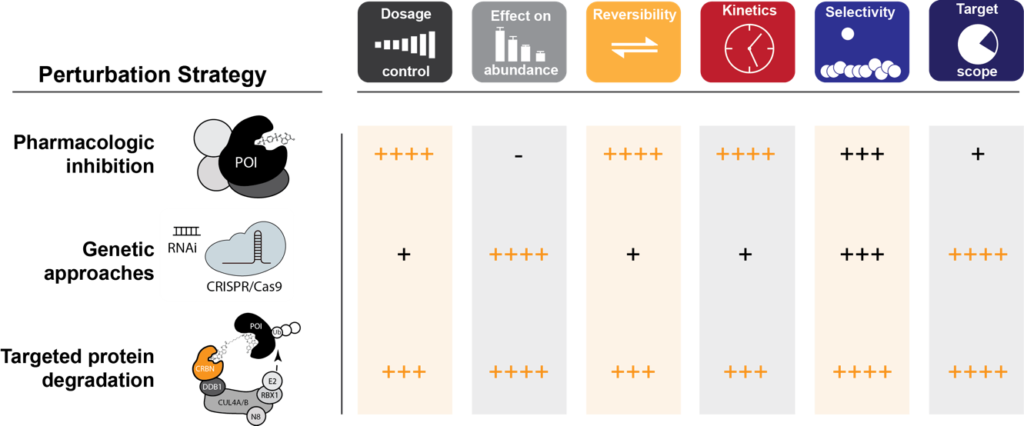
Targeted Protein Degradation as a Validation Strategy. Crucial features are compared among the three key perturbation-approaches in a semi-quantitative manner.
Project members
-
Postdoc
Marko Cigler
SFB Member
Targeted Protein Degradation related publications by Group Winter
- 2024 Degradome analysis to identify direct protein substrates of small-molecule degraders Cell Chemical Biology Go to publication →
- 2024 Inhibitor-induced supercharging of kinase turnover via endogenous proteolytic circuits bioRxiv Go to publication →
- 2024 Monovalent Pseudo-Natural Product Degraders Supercharge the Native Degradation of IDO1 by KLHDC3 bioRxiv Go to publication →
- 2024 Alkylamine-tethered molecules recruit FBXO22 for targeted protein degradation Nature Communication Go to publication →
- 2024 Orpinolide disrupts a leukemic dependency on cholesterol transport by inhibiting OSBP Nature Chemical Biology Go to publication →
- 2024 Large-scale chemoproteomics expedites ligand discovery and predicts ligand behavior in cells Science Go to publication →
- 2024 Targeted protein degradation via intramolecular bivalent glues Nature Go to publication →
- 2023 Discovery of Molecular Glue Degraders via Isogenic Morphological Profiling ASC Chemical Biology Go to publication →
- 2023 Discovery of a Drug-like, Natural Product-Inspired DCAF11 Ligand Chemotype Nature Communication Go to publication →
- 2023 Advancing Targeted Protein Degradation via Multiomics Profiling and Artificial Intelligence Journal of American Chemical Society Go to publication →
- 2023 E3-Specific Degrader Discovery by Dynamic Tracing of Substrate Receptor Abundance Journal of American Chemical Society Go to publication →
- 2022 POLθ processes ssDNA gaps and promotes replication fork progression in BRCA1 deficient cells Cell Reports Go to publication →
- 2022 Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders Nature Chemical Biology Go to publication →
- 2021 Identification and selectivity profiling of small-molecule degraders via multi-omics approaches Cell Chemical Biology Go to publication →