February 10, 2023
Shuffled ATG8 interacting motifs form an ancestral bridge between UFMylation and autophagy
Persistent cellular stress, resulting from disturbances of cellular homeostasis, impairs cell fitness and lifespan. Cellular stress may develop, for example, when ribosomes collide while translating faulty mRNAs. As a result, cells get overburdened with unfinished and improperly formed protein products that form toxic protein aggregates. During cellular stress, cells can call on an arsenal of quality control (QC) mechanisms to restore homeostasis. Cells experiencing prolonged stress in the endoplasmic reticulum (ER), the cellular center for protein synthesis and transport, initiate an ER-specific autophagic pathway called “ER-phagy” to selectively remove damaged ER. When ribosomes collide on the ER, another QC pathway, called “UFMylation”, cooperates with ER-phagy to get rid of incompletely synthesized proteins at the ER membrane. UFMylation is an enigmatic QC pathway based on a protein post-translational modification that resembles ubiquitin and its functions are still being deciphered.
Now, a team of researchers at the Vienna BioCenter, including multiple members of our Special Research Program, uncover an ancient molecular switch that regulates ER-phagy. Using a combination of evolutionary biology and mechanistic experimentation, the researchers demonstrate that the competition between two ubiquitin-like molecules, UFM1 and ATG8 creates a molecular switch in the master regulator C53, thus initiating ER-phagy.
Read the full story in the GMI news here.
Publication:
Shuffled ATG8 interacting motifs form an ancestral bridge between UFMylation and autophagy.
EMBO Journal. 2023 February 10.
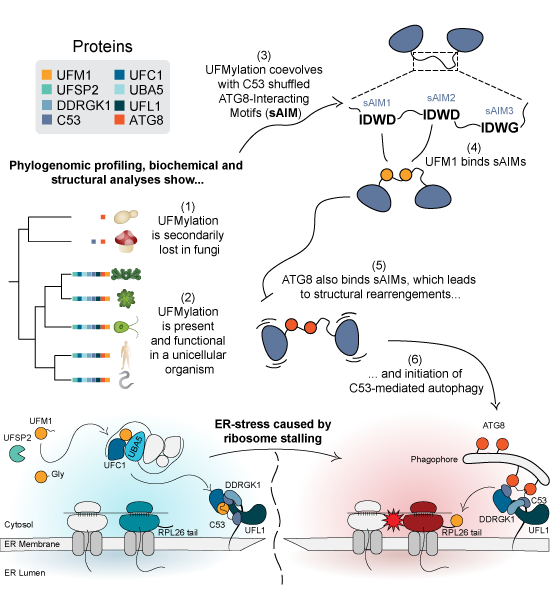
Graphical Synopsis. Phylogenomic profiling has shown that UFMylation coevolves with the shuffled ATG8-interacting motifs (sAIMs) in the ER-phagy receptor C53. Further mechanistic analyses demonstrated that sAIMs regulate C53-mediated autophagy by competitively binding UFM1 and ATG8. ©Picchianti et al., EMBO J. (2023) https://doi.org/10.15252/embj.2022112053