May 22, 2020
“Reverse engineering” autophagosome biogenesis reveals new protein interactions
Scientists from Sascha Martens Lab at the Max Perutz Labs Vienna and their colleagues from the University of Berkeley (USA) have reconstituted the activity of key proteins involved in the growth of autophagosome precursors, a process essential for encapsulating cellular components targeted for degradation and recycling. Their results reveal a previously unknown positive feedback loop and activation mechanism that help explain how the autophagy machinery rapidly generates the autophagosomal membrane. The study is published in the Journal of Cell Biology.
During autophagy, unwanted or dangerous cellular material is captured in vesicles called autophagosomes that deliver the cargo to lysosomes, the waste disposal and recycling centers of the cell. The precursors of autophagosomes are phagophores, small patches of membrane that nucleate the autophagic machinery and engulf the cargo as they grow.
A key protein for phagophore growth and cargo binding is called LC3. LC3 is conjugated to phospholipids on the autophagosomal membrane in a reaction dictated by an enzyme complex (E3). This complex is recruited to the phagophore by sensor proteins (WIPI2) which themselves depend on the activity of a lipid kinase (PI3KC3-C1). The kinase generates a key autophagic signal in the form of another phospholipid (PI3P) that recruits WIPI2 to the pre-autophagosomal membrane. The researchers reconstituted these key steps of membrane targeting and activation in vitro and unravelled how the different “cogwheels” of the autophagy machinery interlock to efficiently generate autophagosomes.
Intriguingly, they showed that the recruitment of PI3KC3-C1 and WIPI2 is mutually enhanced in a positive feedback loop. The more of the kinase was present the more WIPI2 was recruited and vice versa. WIPI2 was also observed to activate the E3 complex at a site other than the active site, a phenomenon known as allosteric activation. “These two effects help explain how the autophagy machine rapidly recruits LC3 and generates the autophagosomal membrane on a timescale similar to that in cells”, says Dorotea Fracchiolla, first author of the study and Postdoc in the Martens lab.
The results are part of a bigger effort between the Martens lab and the Hurley (UC Berkeley), Hummer (MPI, Frankfurt) and Yoshimori (Osaka) labs to reconstitute each step of mammalian autophagosome biogenesis in the lab from start to finish. In live cells, many different processes happen at the same time, making it difficult to observe certain mechanisms in detail. The scientists used purified, recombinant proteins together with giant unilamellar vesicles (GUVs) to characterise individual steps. By systematically adding each component (lipids and proteins) they could study their specific roles in the process, a key advantage over in vivo methods. The research was funded by the Human Frontiers Science Programme (HFSP).
Science Sketch made by Dorotea Fracchiolla:
https://www.youtube.com/watch?v=soWx_tuJm_g
Publication:
Dorotea Fracchiolla, Chunmei Chang, James H. Hurley, Sascha Martens: A PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J Cell Biol (2020) 219 (7): e201912098.
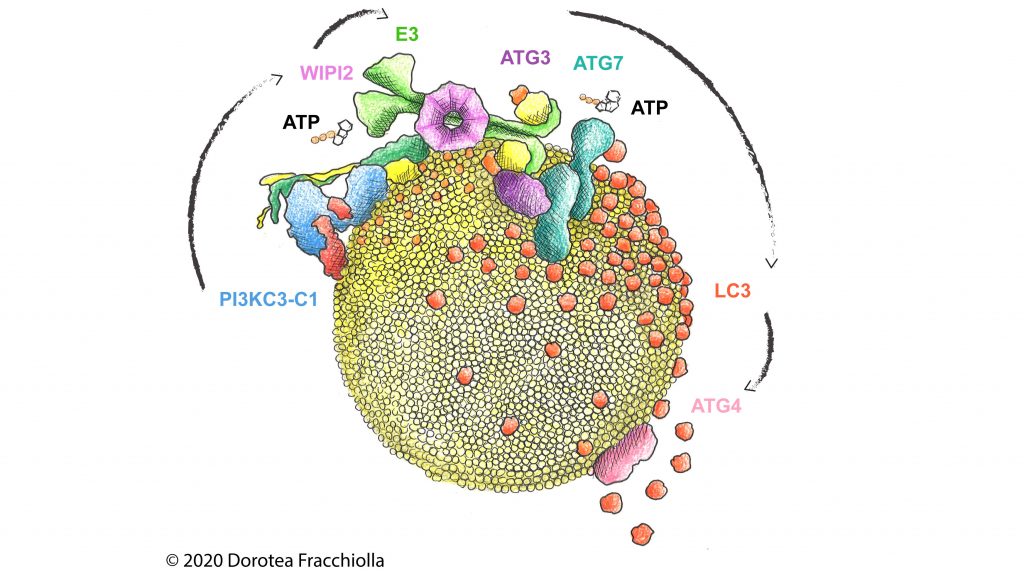
Reconstitution from PI(3)P production to LC3 lipidation on synthetic membranes by the human autophagy machinery. ©Art&Science by Dorotea Fracchiolla www.my-art-science.com