1960s: New puzzle for research
“Self eating” by Cells
Researchers are perplexed about observations of cells self-digesting their own components by engulfing cytoplasmic material within themselves and sending them for lysosomal degradation. The term “Autophagy” is coined by Christian de Duve in 1963.
Image Acknowledgement: Illustration by David S. Goodsell and Daniel Klionsky. doi: 10.2210/rcsb_pdb/goodsell-gallery-012
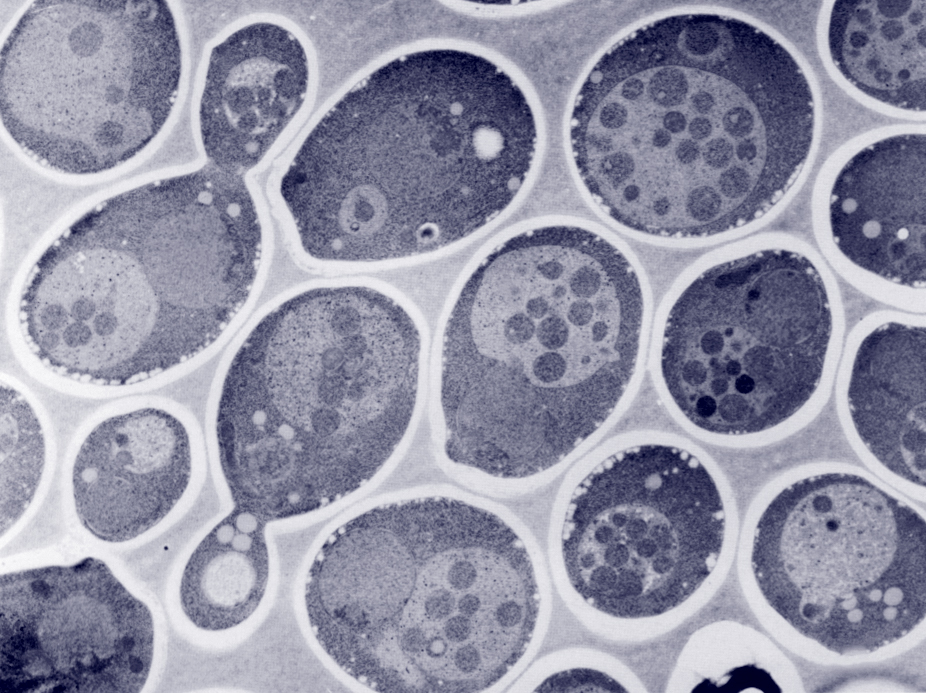
1990s
Discovery of the first autophagy-related genes
The mechanisms underlying autophagy totally lacks understanding until the early 1990s, when Yoshinori Ohsumi isolates the first autophagy-defective mutants, now known as ATG (autophagy-related), in yeast. Identifications of homologous genes in other organisms including mammals follow.
The discovery of the ATG genes triggers an explosive increase in further findings: mechanistic details as well as the physiological relevance of the pathway.
Image Acknowledgement: ©1992 Takeshige K et al. Originally published in Journal of Cell Biology. doi: 10.1083/jcb.119.2.301
2000s
Defects in autophagy cause various diseases
Loss of function analysis of autophagy genes in mice reveal the many roles of autophagy in mammalian differentiation and development, promoting cell survival and organismal health. Mechanistically, discoveries on the role of ubiquitin in autophagy emerge.
Lucas Cranach - Der Jungbrunnen (1546)
2016
Nobel Prize for finding mechanisms of autophagy
Ohsumi's discoveries were critical for our understanding of how the cell degrades and recycles its content. He was awarded the Nobel Prize in Medicine or Physiology in 2016 for his discoveries of mechanisms for autophagy.
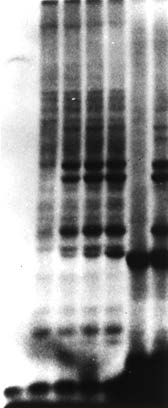
Around 1980
A non-lysosomal degradation pathway exists
Researchers discover a non-lysosomal ATP-dependent proteolytic pathway, and report a heat-stable polypeptide, which later turns out to be ubiquitin, covalently bound to proteins that get degraded.
Image Acknowledgement: Hershko A at al. PNAS USA, 1980. doi: 10.1073/pnas.77.4.1783
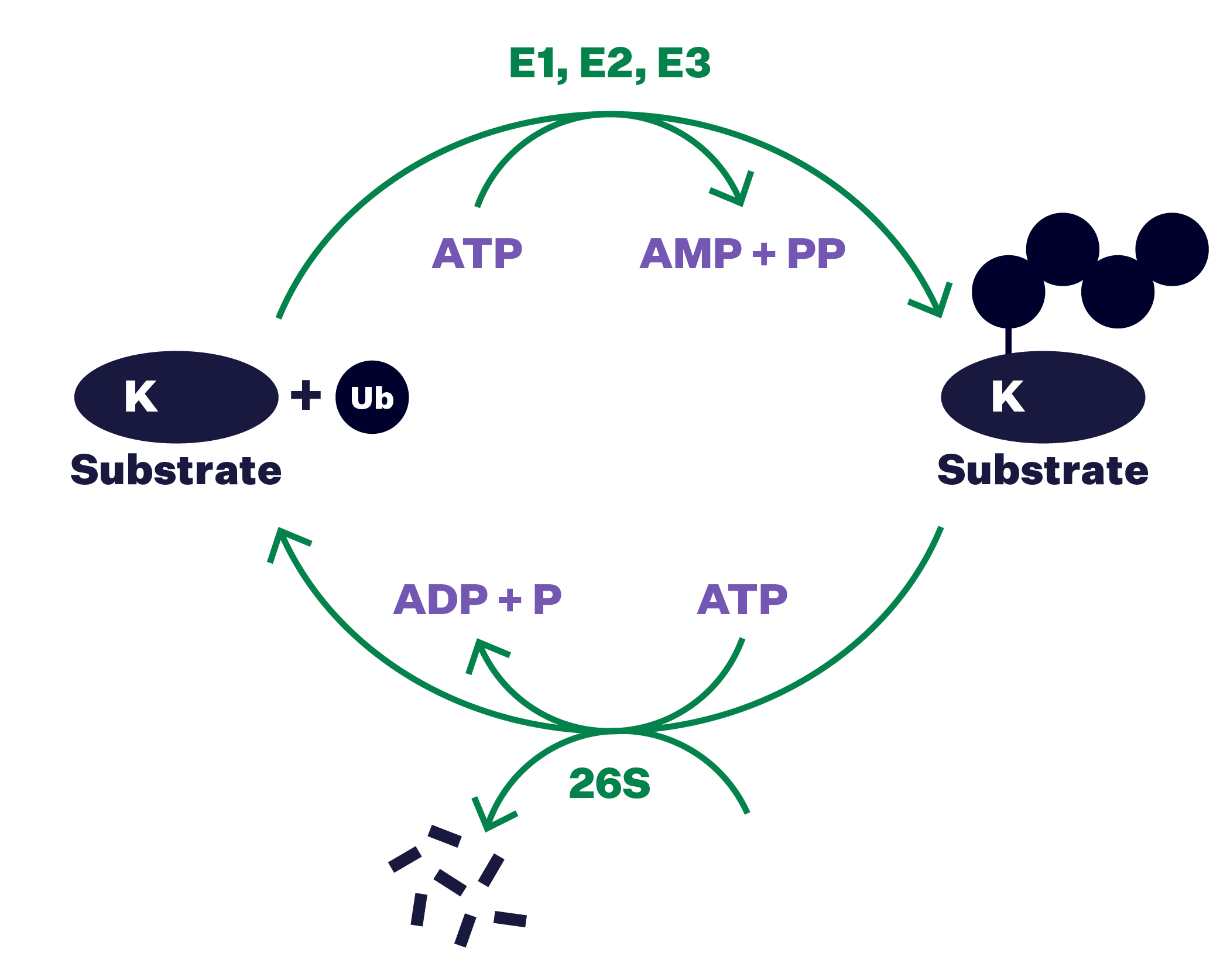
1980—1983
A new protein modification in a new proteolytic pathway
Ubiquitin bound to proteins is demonstrated to be the signal for a downstream yet uncharacterized protease. A cascade of three enzymes, named E1, E2 and E3, is found to mediate ubiquitin conjugation.
1980s & 1990s
Discovery of the proteasome
A multicatalytic protease complex is identified in the late 1980s, followed by the discovery of a larger ATP-dependent protease complex, the 26S protease, that degrades ubiquitin-conjugates. The multicatalytic protease complex is identified as the 20S proteasome, part of the 26S proteasome. The crystal structure of the 20S proteasome is resolved is 1997.

2004
Nobel Prize for the discovery of ubiquitin-mediated protein degradation
Nobel Laureates: Aaron Ciechanover, Avram Herschko, Irwin Rose

2000s
Hijacking the UPS: Make an E3 ligase meet a new substrate
2000s: Researchers prove it possible to target a protein in the cell “at will” to the UPS pathway for degradation using PROTAC, proteolysis targeting chimera: a synthesized bi-functional chimeric molecule that links a desired target protein to a ubiquitin ligase.
Around 2010
Plant hormones are found to bind E3 ligases and enable cognate E3 ligase – substrate pairs to interact. Analogous to this, the concept of „molecular glues“ emerges: synthetic small molecules acting as „molecular glues“ that modify a protein surface to allow and stabilize new E3 ligase – substrate interactions.
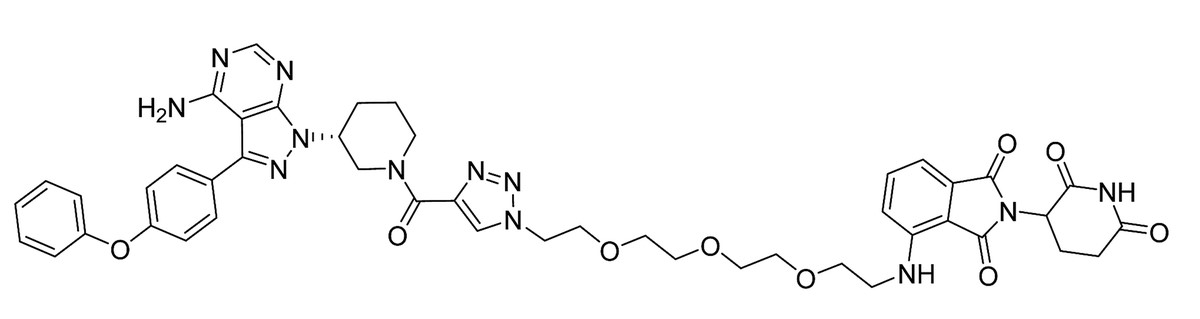
Since 2010
Target “undruggable” proteins by PROTACs or Molecular Glues
The drug thalidomide, previously used without knowing its molecular mechanism of action, is found to act as molecular glue, targeting the E3 ligase cereblon and changing which substrates can be degraded by it.
PROTACs are being developed further: initially peptide-based, now small-molecule PROTACs are reported.
The concept of PROTAC and Molecular Glues has opened a whole new field for targeting disease-causing proteins for destruction. The first PROTAC, an anticancer candidate, reaches clinical trials in 2019.